The iVirus project is a collaboration between the Hurwitz lab, of the University of Arizona, and the Sullivan lab, of the Ohio State University. It arose from noticing the painfully obvious need to make viral ecology tools open and available to the viral ecology community. With the recent (well, for the past decade or so) rise in next-generation sequencing, the field of viral ecology has dramatically expanded our knowledge of the “virosphere”, led in no small part by the dramatically lower cost to sequence environmental viruses. With the dramatic influx of huge amounts of sequencing data, it’s becoming an ever-increasing challenge to process these datasets. Luckily, iVirus leverages the CyVerse cyberinfrastructure, an NSF-funded project that seeks to address the challenges of large-scale data analysis. It comes with data storage (both public and private), a GUI interface, and 1000s of “apps” (read: tools) that researchers can use to process their data. It’s entirely possible to go from raw sequencing data from a sequencing center to fully analyzed viral metagenomes.
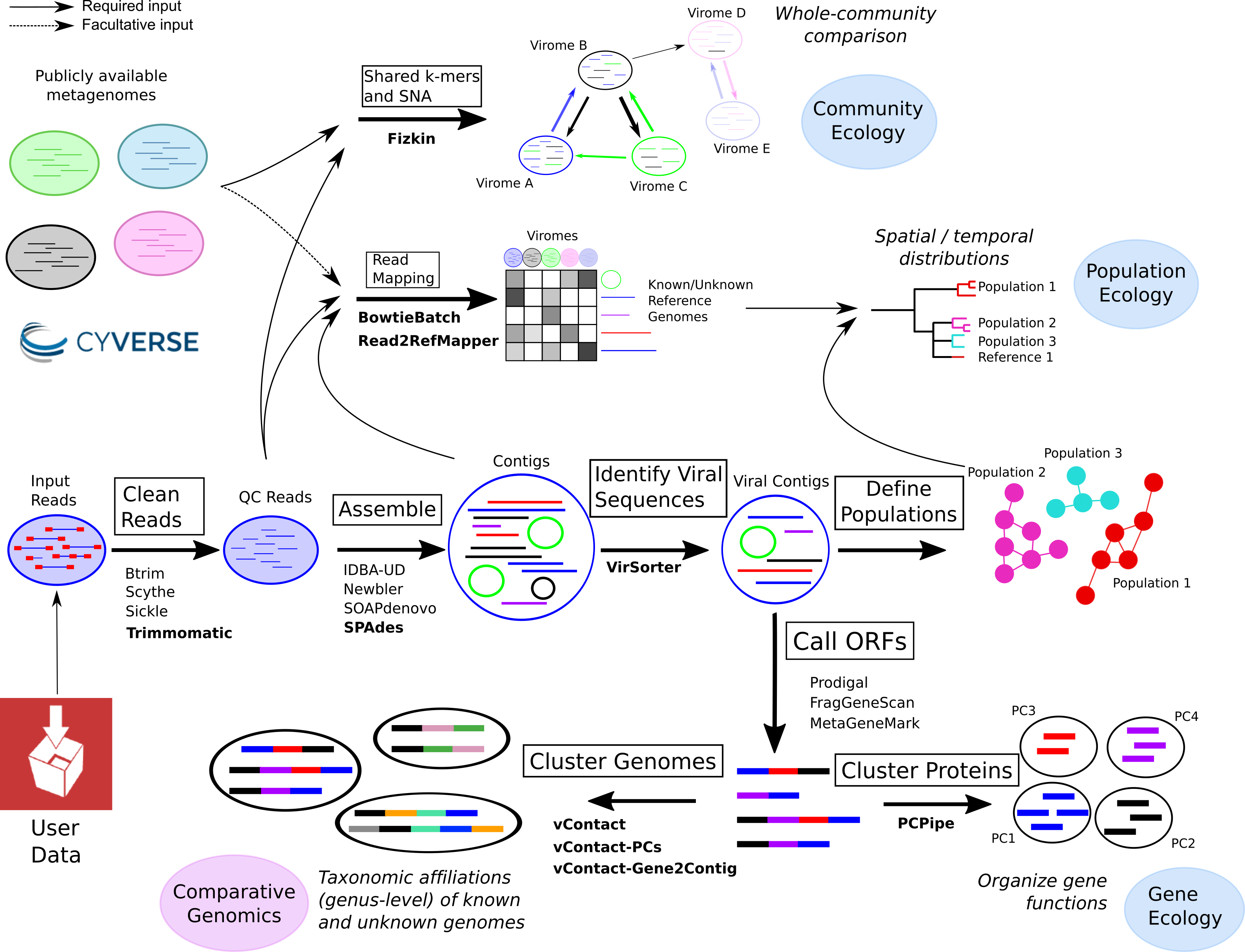
You can read more about the project at its home, the iVirus project.
